We are an interdisciplinary group focussing on the development of algorithms and computational tools to understand, predict and manipulate complex biological networks.
Work in our lab broadly falls under four related themes:
- Understanding microbial interactions in microbiomes
- In silico metabolic engineering
- Theoretical investigations of biological networks
- Biological data analysis
We are also an active member of the Centre for Integrative Biology and Systems mEdicine (IBSE), and the Robert Bosch Centre for Data Sciences and Artificial Intelligence (RBC-DSAI) at IIT Madras.
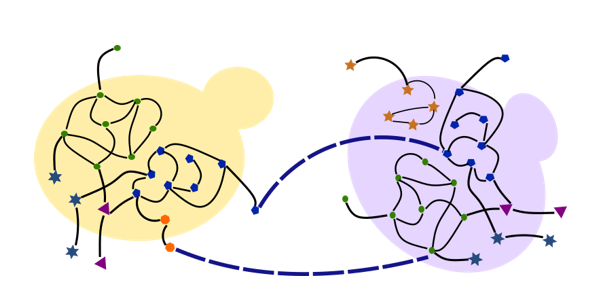
(1) Understanding microbial interactions in microbiomes
Building on our expertise in metabolic modelling, we have now initiated studies of several microbiomes and microbial interactions in these microbiomes. Building on our MetQuest algorithm and Metabolic Support Index to understand microbial metabolic
interactions, we are now studying microbiomes as diverse as the Guaymas Basic hydrothermal vents to the International Space Station microbiome. We are also very much interested in studying the gut microbiome and the role of keystone species in any microbiome.
Projects
- Studying novel microbial species in the ISS
- Studying microbial interactions in the ISS microbiome
- Variations in the ocular microbiome between healthy and keratitis conditions
- Extreme environments such as hydrothermal vents
- Gut microbiome: interactions and organisation
Relevant Publications
- [bibtex file=karthikraman-publications.bib key=Kumar2022Metabolic]
- Metabolic modeling of the International Space Station microbiome reveals key microbial interactions
[bibtex file=karthikraman-publications.bib key=Chng2020Metagenomewide]
[bibtex file=karthikraman-publications.bib key=Ravikrishnan2020Investigating]
[bibtex file=karthikraman-publications.bib key=Ravikrishnan2018Enumerating]
[bibtex file=karthikraman-publications.bib key=Devika2019Deciphering]
(2) In silico Metabolic Engineering
We develop computational approaches to predict ways to manipulate metabolic networks, for the over-production of commercially important molecules, e.g. hyaluronic acid, α-tocopherol. We primarily focus on constraint-based approaches to study metabolic networks, using tools such as flux balance analysis. We also have collaborations with experimental labs, to validate the predicted metabolic engineering strategies.
Projects
- Rational design of a consortium for metabolic engineering
- Overproduction of α-tocopherol in H. annuus cell lines
- Overproduction of hyaluronic acid in L. lactis
- Predicting novel metabolic pathways for retrosynthesis
Relevant Publications
[bibtex file=karthikraman-publications.bib key=Raajaraam2022Computational]
[bibtex file=karthikraman-publications.bib key=Ibrahim2021Twospecies]
[bibtex file=karthikraman-publications.bib key=Ibrahim2021Modelling]
[bibtex file=karthikraman-publications.bib key=Srinivasan2019Rational]
[bibtex file=karthikraman-publications.bib key=Badri2019Uncovering]
[bibtex file=karthikraman-publications.bib key=Sankar2017Predicting]
[bibtex file=karthikraman-publications.bib key=Badri2016In]
[bibtex file=karthikraman-publications.bib key=Ravikrishnan2015Critical]

(3) Theoretical Investigations of Biological Networks
A lot of our work involves a fundamental understanding of biological networks, such as metabolic networks. We try to answer fundamental questions about the organisation and evolution of these networks, in a bid to better understand the constraints that permeate their design. This, in turn, will be very helpful towards designing and manipulating such networks, for various applications, including metabolic engineering.
Projects
- Understanding synthetic lethality in metabolic networks
- Robustness and plasticity of metabolic networks
- Design principles of oscillators, network design for synthetic biology
- Systems-theoretic approaches to design adaptive networks
- Sloppiness in biological systems
Relevant Publications
[bibtex file=karthikraman-publications.bib key=Bhattacharya2022Discovering]
[bibtex file=karthikraman-publications.bib key=Sambamoorthy2020MinReact]
[bibtex file=karthikraman-publications.bib key=Bhattacharya2021SystemsTheoretic]
[bibtex file=karthikraman-publications.bib key=bhattacharya2018systemstheoretic]
[bibtex file=karthikraman-publications.bib key=Sambamoorthy2019Evolutionary]
[bibtex file=karthikraman-publications.bib key=Sambamoorthy2018Understanding]
[bibtex file=karthikraman-publications.bib key=Pratapa2015FastSL]
[bibtex file=karthikraman-publications.bib key=Partha2014Revisiting]
[bibtex file=karthikraman-publications.bib key=Raman2014Organisational]
(4) Biological Data Analysis
We are also working on a data-centric investigation of biological networks. For example, how can we understand biological networks, by using multi-dimensional data (e.g. genomic, transcriptomic, proteomic, phosphoproteomic etc.)? We also apply techniques from machine learning to study biological networks and datasets alike, to make testable predictions and generate hypotheses for wet lab experimentation.
Projects
- Predicting essential proteins in protein interaction networks (See NetGenes database)
- Identifying disease modules in biological networks [DREAM Challenge]
- Identifying the context of mutations in cancer (DBT Project)
Relevant Publications
[bibtex file=karthikraman-publications.bib key=Sudhakar2022Multiomic]
[bibtex file=karthikraman-publications.bib key=Sudhakar2022Novel]
[bibtex file=karthikraman-publications.bib key=Senthamizhan2021NetGenes]
[bibtex file=karthikraman-publications.bib key=Banerjee2021Sequence]
[bibtex file=karthikraman-publications.bib key=Choobdar2019Assessment]
[bibtex file=karthikraman-publications.bib key=Tripathi2019adapting]
[bibtex file=karthikraman-publications.bib key=azhagesan2018networkbased]

