Background
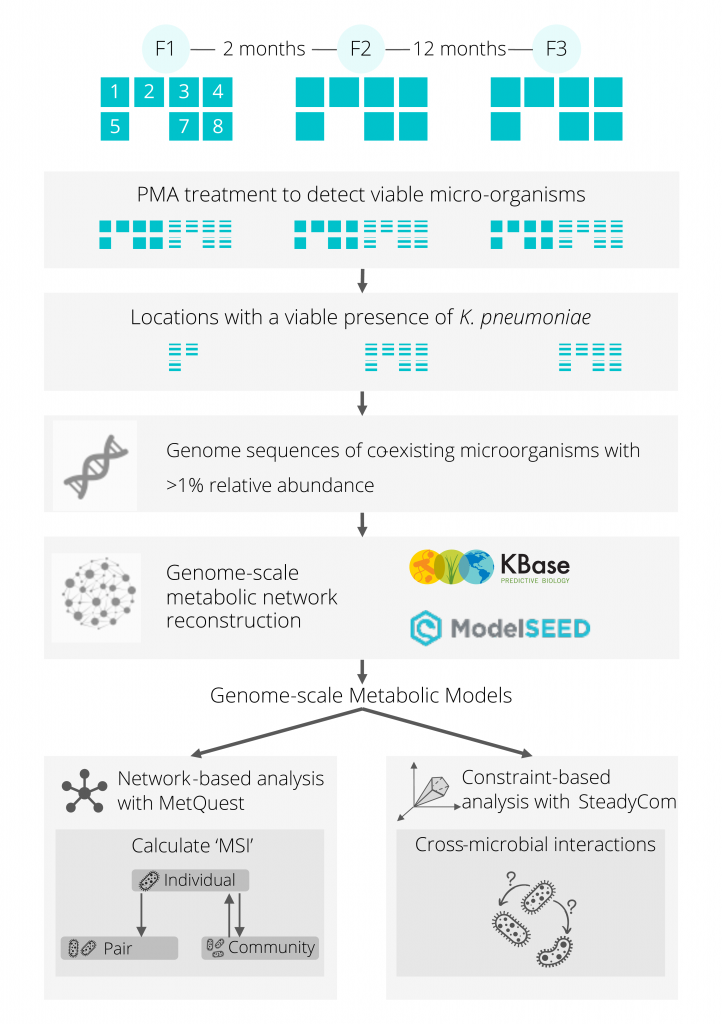
Recent studies have provided insights into the persistence and succession of microbes aboard the International Space Station (ISS), notably the dominance of Klebsiella pneumoniae. However, the interactions between the various microbes aboard the ISS, and how it shapes the microbiome remain to be clearly understood. In this study, we apply a computational approach to predict possible metabolic interactions in the ISS microbiome and shed further light on its organization.
Results
Through a combination of a systems-based graph-theoretical approach, and a constraint-based community metabolic modelling approach, we demonstrated several key interactions in the ISS microbiome. These complementary approaches provided insights into the metabolic interactions and dependencies present amongst various microbes in a community, highlighting key interactions and keystone species. Our results showed that the presence of K. pneumoniae is beneficial to many other microorganisms it coexists with, notably those from the Pantoea genus. Species belonging to the Enterobacteriaceae family were often found to be the most beneficial for the survival of other microorganisms in the ISS microbiome. However, K. pneumoniae was found to exhibit parasitic and amensalistic interactions with Aspergillus and Penicillium species, respectively. To prove this metabolic prediction, K. pneumoniae and Aspergillus fumigatus were co-cultured under normal and simulated microgravity, where K. pneumoniae cells showed parasitic characteristics to the fungus. The electron micrography revealed that the presence of K. pneumoniae compromised the morphology of fungal conidia and degenerated its biofilm-forming structures.
Conclusion
Our study underscores the importance of K. pneumoniae in the ISS, and its potential positive and negative interactions with other microbes, including potential pathogens. This integrated modelling approach, combined with experiments, demonstrates potential for understanding the organization of other such microbiomes, unravelling key organisms and their interdependencies.
Original Paper: [bibtex file=karthikraman-publications.bib key=Kumar2022Metabolic]

